Variable selection using genetic algorithm
Background
Genetic algorithm is inspired by a natural selection process by which the fittest individuals be selected to reproduce. This algorithm has been used in optimization and search problem, and also, can be used for variable selection.
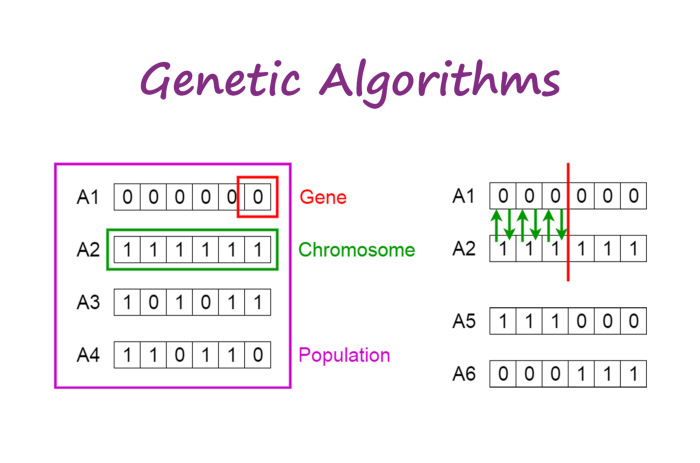
Genetic algorithm - gene, chromosome, population, crossover (upper right), offspring (lower right)
First, let’s go into a few terms related to genetic algorithm theory.
- Population - a set of chromosomes
- Chromosome - a subset of variables (also known as individual by some reference)
- Gene - a variable or feature
- Fitness function - give fitness score to each chromosome and guide the selection
- Selection - a process to select the two chromosome known as parents
- Crossover - a process to generate offspring by parents (illustrate in the picture above, on the upper right side)
- Mutation - the process by which the gene in the chromosome is randomly flipped into 1 or 0

Mutation
So, the basic flow of genetic algorithm:
Algorithm starts with an initial population, often randomly generated
Create a successive generation by selecting a portion of the initial population (the selection is guided by the fitness function) - this includes selection -> crossover -> mutation
The algorithm terminates if certain predetermined criteria are met such as:
- Solution satisfies the minimum criteria
- Fixed number of generation reached
- Successive iteration no longer produce a better result
- Solution satisfies the minimum criteria
Example in R
There is GA package in R, where we can implement the genetic algorithm a bit more manually where we can specify our own fitness function. However, I think it is easier to use a genetic algorithm implemented in caret package for variable selection.
Load the packages.
library(caret)
library(tidyverse)
library(rsample)
library(recipes)The data.
dat <-
mtcars %>%
mutate(across(c(vs, am), as.factor),
am = fct_recode(am, auto = "0", man = "1"))
str(dat)## 'data.frame': 32 obs. of 11 variables:
## $ mpg : num 21 21 22.8 21.4 18.7 18.1 14.3 24.4 22.8 19.2 ...
## $ cyl : num 6 6 4 6 8 6 8 4 4 6 ...
## $ disp: num 160 160 108 258 360 ...
## $ hp : num 110 110 93 110 175 105 245 62 95 123 ...
## $ drat: num 3.9 3.9 3.85 3.08 3.15 2.76 3.21 3.69 3.92 3.92 ...
## $ wt : num 2.62 2.88 2.32 3.21 3.44 ...
## $ qsec: num 16.5 17 18.6 19.4 17 ...
## $ vs : Factor w/ 2 levels "0","1": 1 1 2 2 1 2 1 2 2 2 ...
## $ am : Factor w/ 2 levels "auto","man": 2 2 2 1 1 1 1 1 1 1 ...
## $ gear: num 4 4 4 3 3 3 3 4 4 4 ...
## $ carb: num 4 4 1 1 2 1 4 2 2 4 ...For this, we going to use random forest (rfGA). Other options are bagged tree (treebagGA) and caretGA. We are able to use other method in caret if we use caretGA.
# specify control
ga_ctrl <- gafsControl(functions = rfGA, method = "cv", number = 5)
# run random forest
set.seed(123)
rf_ga <- gafs(x = dat %>% select(-am),
y = dat$am,
iters = 5,
gafsControl = ga_ctrl)
rf_ga##
## Genetic Algorithm Feature Selection
##
## 32 samples
## 10 predictors
## 2 classes: 'auto', 'man'
##
## Maximum generations: 5
## Population per generation: 50
## Crossover probability: 0.8
## Mutation probability: 0.1
## Elitism: 0
##
## Internal performance values: Accuracy, Kappa
## Subset selection driven to maximize internal Accuracy
##
## External performance values: Accuracy, Kappa
## Best iteration chose by maximizing external Accuracy
## External resampling method: Cross-Validated (5 fold)
##
## During resampling:
## * the top 5 selected variables (out of a possible 10):
## qsec (60%), wt (60%), disp (40%), gear (40%), vs (40%)
## * on average, 3.2 variables were selected (min = 1, max = 7)
##
## In the final search using the entire training set:
## * 7 features selected at iteration 3 including:
## cyl, hp, drat, qsec, vs ...
## * external performance at this iteration is
##
## Accuracy Kappa
## 0.9429 0.8831The optimal features/variables:
rf_ga$optVariables## [1] "cyl" "hp" "drat" "qsec" "vs" "gear" "carb"This is the time taken for random forest approach.
rf_ga$times## $everything
## user system elapsed
## 51.22 1.25 52.92By default the algorithm will find a solution or a set of variable that reduce RMSE for numerical outcome, and accuracy for categorical outcome. Also, genetic algorithm tend to overfit, that’s why for the implementation in caret we have internal and external performance. So, for the 10-fold cross-validation, 10 genetic algorithm will be run separately. All the first nine folds are used for the genetic algorithm, and the 10th for external performance evaluation.
Let’s try a variable selection using linear regression model.
# specify control
lm_ga_ctrl <- gafsControl(functions = caretGA, method = "cv", number = 5)
# run lm
set.seed(123)
lm_ga <- gafs(x = dat %>% select(-mpg),
y = dat$mpg,
iters = 5,
gafsControl = lm_ga_ctrl,
# below is the option for `train`
method = "lm",
trControl = trainControl(method = "cv", allowParallel = F))
lm_ga##
## Genetic Algorithm Feature Selection
##
## 32 samples
## 10 predictors
##
## Maximum generations: 5
## Population per generation: 50
## Crossover probability: 0.8
## Mutation probability: 0.1
## Elitism: 0
##
## Internal performance values: RMSE, Rsquared, MAE
## Subset selection driven to minimize internal RMSE
##
## External performance values: RMSE, Rsquared, MAE
## Best iteration chose by minimizing external RMSE
## External resampling method: Cross-Validated (5 fold)
##
## During resampling:
## * the top 5 selected variables (out of a possible 10):
## wt (100%), hp (80%), carb (60%), cyl (60%), am (40%)
## * on average, 4.4 variables were selected (min = 4, max = 5)
##
## In the final search using the entire training set:
## * 5 features selected at iteration 5 including:
## cyl, disp, hp, wt, qsec
## * external performance at this iteration is
##
## RMSE Rsquared MAE
## 3.3434 0.7624 2.6037Now, let’s see how to integrate this in machine learning flow using recipes from rsample.
First, we split the data.
set.seed(123)
dat_split <-initial_split(dat)
dat_train <- training(dat_split)
dat_test <- testing(dat_split)We specify two recipes for numerical and categorical outcome.
# Numerical
rec_num <-
recipe(mpg ~., data = dat_train) %>%
step_center(all_numeric()) %>%
step_dummy(all_nominal_predictors())
# Categorical
rec_cat <-
recipe(am ~., data = dat_train) %>%
step_center(all_numeric()) %>%
step_dummy(all_nominal_predictors())We run random forest for numerical outcome recipes.
# specify control
rf_ga_ctrl <- gafsControl(functions = rfGA, method = "cv", number = 5)
# run random forest
set.seed(123)
rf_ga2 <-
gafs(rec_num,
data = dat_train,
iters = 5,
gafsControl = rf_ga_ctrl)
rf_ga2##
## Genetic Algorithm Feature Selection
##
## 24 samples
## 10 predictors
##
## Maximum generations: 5
## Population per generation: 50
## Crossover probability: 0.8
## Mutation probability: 0.1
## Elitism: 0
##
## Internal performance values: RMSE, Rsquared
## Subset selection driven to minimize internal RMSE
##
## External performance values: RMSE, Rsquared, MAE
## Best iteration chose by minimizing external RMSE
## External resampling method: Cross-Validated (5 fold)
##
## During resampling:
## * the top 5 selected variables (out of a possible 10):
## cyl (80%), disp (80%), hp (80%), wt (80%), carb (60%)
## * on average, 4.8 variables were selected (min = 2, max = 9)
##
## In the final search using the entire training set:
## * 6 features selected at iteration 5 including:
## cyl, disp, hp, wt, gear ...
## * external performance at this iteration is
##
## RMSE Rsquared MAE
## 2.830 0.928 2.408The optimal variables.
rf_ga2$optVariables## [1] "cyl" "disp" "hp" "wt" "gear" "vs_X1"Let’s try run SVM for the numerical outcome recipes.
# specify control
svm_ga_ctrl <- gafsControl(functions = caretGA, method = "cv", number = 5)
# run SVM
set.seed(123)
svm_ga <-
gafs(rec_cat,
data = dat_train,
iters = 5,
gafsControl = svm_ga_ctrl,
# below is the options to `train` for caretGA
method = "svmRadial", #SVM with Radial Basis Function Kernel
trControl = trainControl(method = "cv", allowParallel = T))
svm_ga##
## Genetic Algorithm Feature Selection
##
## 24 samples
## 10 predictors
## 2 classes: 'auto', 'man'
##
## Maximum generations: 5
## Population per generation: 50
## Crossover probability: 0.8
## Mutation probability: 0.1
## Elitism: 0
##
## Internal performance values: Accuracy, Kappa
## Subset selection driven to maximize internal Accuracy
##
## External performance values: Accuracy, Kappa
## Best iteration chose by maximizing external Accuracy
## External resampling method: Cross-Validated (5 fold)
##
## During resampling:
## * the top 5 selected variables (out of a possible 10):
## wt (80%), qsec (60%), vs_X1 (60%), carb (40%), disp (40%)
## * on average, 4 variables were selected (min = 3, max = 6)
##
## In the final search using the entire training set:
## * 9 features selected at iteration 2 including:
## mpg, cyl, disp, hp, drat ...
## * external performance at this iteration is
##
## Accuracy Kappa
## 0.9200 0.8571The optimal variables.
svm_ga$optVariables## [1] "mpg" "cyl" "disp" "hp" "drat" "wt" "qsec" "carb" "vs_X1"Conclusion
Although genetic algorithm seems quite good for variable selection, the main limitation I would say is the computational time. However, if we have a lot of variables or features to reduced, using the genetic algorithm despite the long computational time seems beneficial to me.
Reference: