Using UMAP preprocessing for image classification
UMAP
Uniform manifold approximation and projection or in short UMAP is a type of dimension reduction techniques. So, basically UMAP will project a set of features into a smaller space. UMAP can be a supervised technique in which we give a label or an outcome or an unsupervised one. For those interested to know in detail how UMAP works can refer to this reference. For those prefer a much simpler or shorter version of it, I recommend a YouTube video by Joshua Starmer.
Example in R
We going to see how to apply a UMAP techniques for image preprocessing and further classify the images using kNN and naive bayes.
These are the packages that we need.
library(keras) #for data and reshape to tabular format
library(tidymodels)
library(embed) #for umap
library(discrim) #for naive bayes modelWe going to use the famous MNIST dataset. This dataset contained a handwritten digit from 0 to 9. This dataset is available in keras package.
mnist_data <- dataset_mnist()## Loaded Tensorflow version 2.2.0image_data <- mnist_data$train$x
image_labels <- mnist_data$train$y
image_data %>% dim()## [1] 60000 28 28For example this is the image for the second row.
image_data[2, 1:28, 1:28] %>%
t() %>%
image(col = gray.colors(256))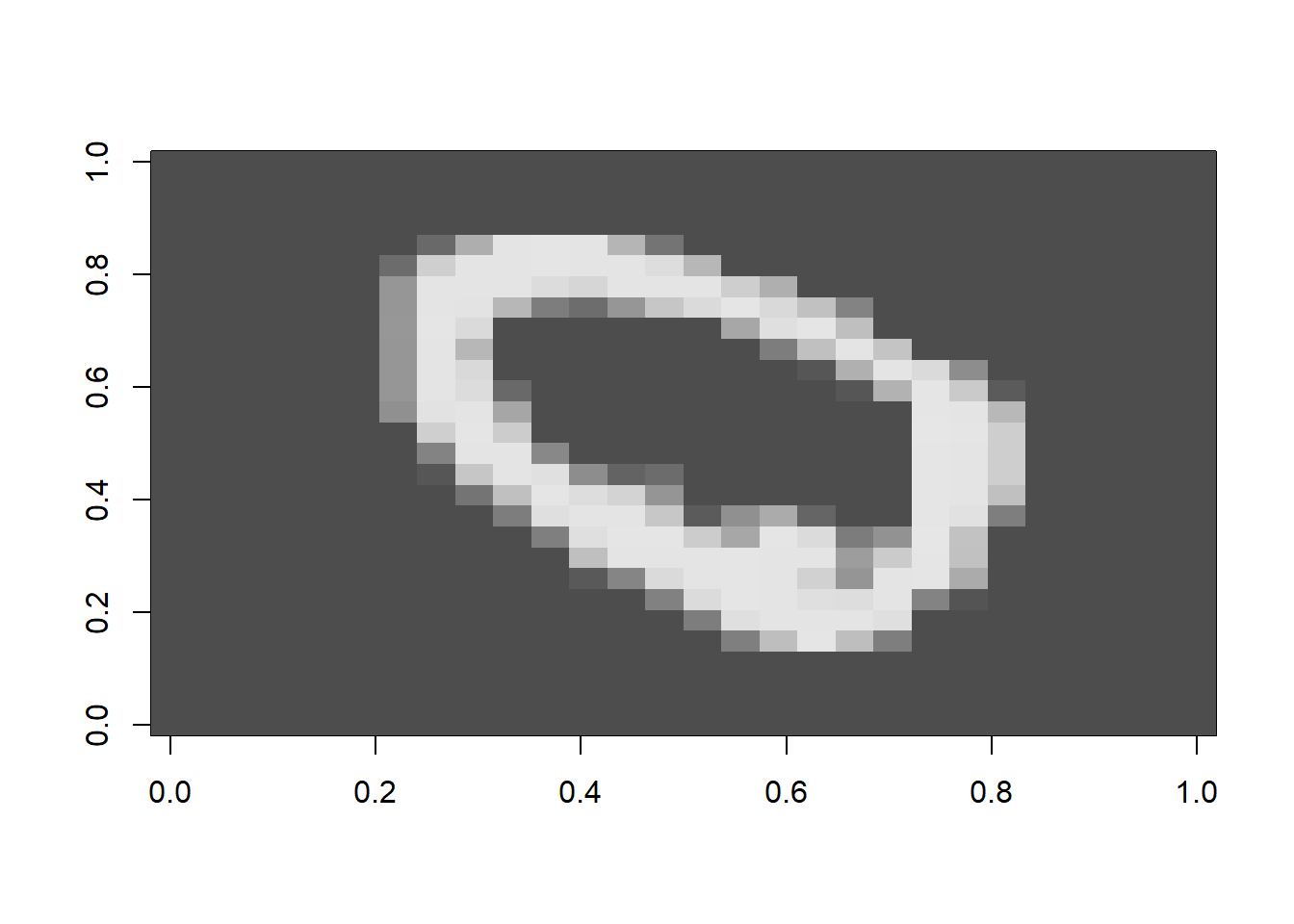
Next, we going to change the image into a tabular data frame format. We going to limit the data to the first 1000 rows or images out of the total 6000 images.
# Reformat to tabular format
image_data <- array_reshape(image_data, dim = c(60000, 28*28))
image_data %>% dim()## [1] 60000 784image_data <- image_data[1:10000,]
image_labels <- image_labels[1:10000]
# Reformat to data frame
full_data <-
data.frame(image_data) %>%
bind_cols(label = image_labels) %>%
mutate(label = as.factor(label))Then, we going to split the data and create a 3-folds cross-validation sets for the sake of simplicity.
# Split data
set.seed(123)
ind <- initial_split(full_data)
data_train <- training(ind)
data_test <- testing(ind)
# 10-folds CV
set.seed(123)
data_cv <- vfold_cv(data_train, v = 3)For recipe specification, we going to scale and center all the predictor after creating a new variable using step_umap(). Notice that in step_umap() we supply the outcome and we tune the number of components (num_comp).
rec <-
recipe(label ~ ., data = data_train) %>%
step_umap(all_predictors(), outcome = vars(label), num_comp = tune()) %>%
step_center(all_predictors()) %>%
step_scale(all_predictors())We create a a base workflow.
wf <-
workflow() %>%
add_recipe(rec) We going to use two models as classifier:
- kNN
- Naive bayes
For each classifier, we going to create a regular grid of parameters to be tuned and further run a regular grid search.
For kNN.
# knn model
knn_mod <-
nearest_neighbor(neighbors = tune()) %>%
set_mode("classification") %>%
set_engine("kknn")
# knn grid
knn_grid <- grid_regular(neighbors(), num_comp(range = c(2, 8)), levels = 3)
# Tune grid search
knn_tune <-
tune_grid(
wf %>% add_model(knn_mod),
resamples = data_cv,
grid = knn_grid,
control = control_grid(verbose = F)
)For naive bayes.
# nb model
nb_mod <-
naive_Bayes(smoothness = tune()) %>%
set_mode("classification") %>%
set_engine("naivebayes")
# nb grid
nb_grid <- grid_regular(smoothness(), num_comp(range = c(2, 10)), levels = 3)
# Tune grid search
nb_tune <-
tune_grid(
wf %>% add_model(nb_mod),
resamples = data_cv,
grid = nb_grid,
control = control_grid(verbose = F)
)Let’s see our tuning performance of our model.
# knn model
knn_tune %>%
show_best("roc_auc")## # A tibble: 5 x 8
## neighbors num_comp .metric .estimator mean n std_err .config
## <int> <int> <chr> <chr> <dbl> <int> <dbl> <chr>
## 1 10 8 roc_auc hand_till 0.961 3 0.000268 Preprocessor3_Mode~
## 2 10 5 roc_auc hand_till 0.961 3 0.000421 Preprocessor2_Mode~
## 3 5 8 roc_auc hand_till 0.959 3 0.000757 Preprocessor3_Mode~
## 4 10 2 roc_auc hand_till 0.959 3 0.000737 Preprocessor1_Mode~
## 5 5 5 roc_auc hand_till 0.958 3 0.000740 Preprocessor2_Mode~knn_tune %>%
show_best("accuracy")## # A tibble: 5 x 8
## neighbors num_comp .metric .estimator mean n std_err .config
## <int> <int> <chr> <chr> <dbl> <int> <dbl> <chr>
## 1 10 8 accuracy multiclass 0.914 3 0.00104 Preprocessor3_Mode~
## 2 5 8 accuracy multiclass 0.913 3 0.00315 Preprocessor3_Mode~
## 3 10 5 accuracy multiclass 0.912 3 0.00114 Preprocessor2_Mode~
## 4 5 5 accuracy multiclass 0.91 3 0.00139 Preprocessor2_Mode~
## 5 10 2 accuracy multiclass 0.910 3 0.00175 Preprocessor1_Mode~# nb model
nb_tune %>%
show_best("roc_auc")## # A tibble: 5 x 8
## smoothness num_comp .metric .estimator mean n std_err .config
## <dbl> <int> <chr> <chr> <dbl> <int> <dbl> <chr>
## 1 1.5 10 roc_auc hand_till 0.971 3 0.000400 Preprocessor3_Mod~
## 2 1.5 6 roc_auc hand_till 0.971 3 0.000997 Preprocessor2_Mod~
## 3 1 10 roc_auc hand_till 0.971 3 0.000634 Preprocessor3_Mod~
## 4 1 6 roc_auc hand_till 0.970 3 0.00124 Preprocessor2_Mod~
## 5 0.5 10 roc_auc hand_till 0.969 3 0.000808 Preprocessor3_Mod~nb_tune %>%
show_best("accuracy")## # A tibble: 5 x 8
## smoothness num_comp .metric .estimator mean n std_err .config
## <dbl> <int> <chr> <chr> <dbl> <int> <dbl> <chr>
## 1 1 10 accuracy multiclass 0.913 3 0.000481 Preprocessor3_Mo~
## 2 1.5 10 accuracy multiclass 0.913 3 0.000267 Preprocessor3_Mo~
## 3 0.5 10 accuracy multiclass 0.912 3 0.000462 Preprocessor3_Mo~
## 4 1.5 6 accuracy multiclass 0.911 3 0.00135 Preprocessor2_Mo~
## 5 1 6 accuracy multiclass 0.910 3 0.00157 Preprocessor2_Mo~Next, we going to select the best model from the tuned parameters and finalise our model using last_fit().
For knn model.
# Finalize
knn_best <- knn_tune %>% select_best("roc_auc")
knn_rec <-
recipe(label ~ ., data = data_train) %>%
step_umap(all_predictors(), outcome = vars(label), num_comp = knn_best$num_comp) %>%
step_center(all_predictors()) %>%
step_scale(all_predictors())
knn_wf <-
workflow() %>%
add_recipe(knn_rec) %>%
add_model(knn_mod) %>%
finalize_workflow(knn_best)
# Last fit
knn_lastfit <-
knn_wf %>%
last_fit(ind)For naive bayes model.
# Finalize
nb_best <- nb_tune %>% select_best("roc_auc")
nb_rec <-
recipe(label ~ ., data = data_train) %>%
step_umap(all_predictors(), outcome = vars(label), num_comp = nb_best$num_comp) %>%
step_center(all_predictors()) %>%
step_scale(all_predictors())
nb_wf <-
workflow() %>%
add_recipe(nb_rec) %>%
add_model(nb_mod) %>%
finalize_workflow(nb_best)
# Last fit
nb_lastfit <-
nb_wf %>%
last_fit(ind)Let’s see the model performance on the testing data.
knn_lastfit %>%
collect_metrics() %>%
mutate(model = "knn") %>%
dplyr::bind_rows(nb_lastfit %>%
collect_metrics() %>%
mutate(model = "nb")) %>%
select(-.config)## # A tibble: 4 x 4
## .metric .estimator .estimate model
## <chr> <chr> <dbl> <chr>
## 1 accuracy multiclass 0.938 knn
## 2 roc_auc hand_till 0.971 knn
## 3 accuracy multiclass 0.936 nb
## 4 roc_auc hand_till 0.980 nbThese are the confusion matrices.
knn_lastfit %>%
collect_predictions() %>%
conf_mat(label, .pred_class) %>%
autoplot(type = "heatmap") +
labs(title = "Confusion matrix - kNN")
nb_lastfit %>%
collect_predictions() %>%
conf_mat(label, .pred_class) %>%
autoplot(type = "heatmap") +
labs(title = "Confusion matrix - naive bayes")
Lastly, we can compare the ROC plots for each class.
knn_lastfit %>%
collect_predictions() %>%
mutate(id = "knn") %>%
bind_rows(
nb_lastfit %>%
collect_predictions() %>%
mutate(id = "nb")
) %>%
group_by(id) %>%
roc_curve(label, .pred_0:.pred_9) %>%
autoplot()
Conclusion
I believe UMAP is quite good and can be used as one of preprocessing step in image classification. We are able to get a pretty good performance result in this post. I believe if the the parameter tuning approach is a bit more rigorous, the performance result will be a lot better.