A summary of forcats package

I just watched a youtube video by Andrew Couch about his commonly used function in readr, stringr, and forcats packages. Although, I have used forcats package before, I realised that I have not fully utilised all of its function.
So, in this post, I have summarised main function of forcats that I find useful in my day-to-day R coding. Basically, more like a note to myself.
Main functions
We will use mtcars data to demonstrate each function. forcats is part of tiyverse packages. So, it will load, once we load the tidyverse packages.
library(tidyverse)
glimpse(mtcars)## Rows: 32
## Columns: 11
## $ mpg <dbl> 21.0, 21.0, 22.8, 21.4, 18.7, 18.1, 14.3, 24.4, 22.8, 19.2, 17.8,~
## $ cyl <dbl> 6, 6, 4, 6, 8, 6, 8, 4, 4, 6, 6, 8, 8, 8, 8, 8, 8, 4, 4, 4, 4, 8,~
## $ disp <dbl> 160.0, 160.0, 108.0, 258.0, 360.0, 225.0, 360.0, 146.7, 140.8, 16~
## $ hp <dbl> 110, 110, 93, 110, 175, 105, 245, 62, 95, 123, 123, 180, 180, 180~
## $ drat <dbl> 3.90, 3.90, 3.85, 3.08, 3.15, 2.76, 3.21, 3.69, 3.92, 3.92, 3.92,~
## $ wt <dbl> 2.620, 2.875, 2.320, 3.215, 3.440, 3.460, 3.570, 3.190, 3.150, 3.~
## $ qsec <dbl> 16.46, 17.02, 18.61, 19.44, 17.02, 20.22, 15.84, 20.00, 22.90, 18~
## $ vs <dbl> 0, 0, 1, 1, 0, 1, 0, 1, 1, 1, 1, 0, 0, 0, 0, 0, 0, 1, 1, 1, 1, 0,~
## $ am <dbl> 1, 1, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1, 1, 0, 0,~
## $ gear <dbl> 4, 4, 4, 3, 3, 3, 3, 4, 4, 4, 4, 3, 3, 3, 3, 3, 3, 4, 4, 4, 3, 3,~
## $ carb <dbl> 4, 4, 1, 1, 2, 1, 4, 2, 2, 4, 4, 3, 3, 3, 4, 4, 4, 1, 2, 1, 1, 2,~There are 9 forcats functions that I think very useful.
factor()
factor() changes variable type into a factor or categorical type
mtcars$carb <- factor(mtcars$carb)
glimpse(mtcars)## Rows: 32
## Columns: 11
## $ mpg <dbl> 21.0, 21.0, 22.8, 21.4, 18.7, 18.1, 14.3, 24.4, 22.8, 19.2, 17.8,~
## $ cyl <dbl> 6, 6, 4, 6, 8, 6, 8, 4, 4, 6, 6, 8, 8, 8, 8, 8, 8, 4, 4, 4, 4, 8,~
## $ disp <dbl> 160.0, 160.0, 108.0, 258.0, 360.0, 225.0, 360.0, 146.7, 140.8, 16~
## $ hp <dbl> 110, 110, 93, 110, 175, 105, 245, 62, 95, 123, 123, 180, 180, 180~
## $ drat <dbl> 3.90, 3.90, 3.85, 3.08, 3.15, 2.76, 3.21, 3.69, 3.92, 3.92, 3.92,~
## $ wt <dbl> 2.620, 2.875, 2.320, 3.215, 3.440, 3.460, 3.570, 3.190, 3.150, 3.~
## $ qsec <dbl> 16.46, 17.02, 18.61, 19.44, 17.02, 20.22, 15.84, 20.00, 22.90, 18~
## $ vs <dbl> 0, 0, 1, 1, 0, 1, 0, 1, 1, 1, 1, 0, 0, 0, 0, 0, 0, 1, 1, 1, 1, 0,~
## $ am <dbl> 1, 1, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1, 1, 0, 0,~
## $ gear <dbl> 4, 4, 4, 3, 3, 3, 3, 4, 4, 4, 4, 3, 3, 3, 3, 3, 3, 4, 4, 4, 3, 3,~
## $ carb <fct> 4, 4, 1, 1, 2, 1, 4, 2, 2, 4, 4, 3, 3, 3, 4, 4, 4, 1, 2, 1, 1, 2,~fct_inorder()
This function sorts factor levels based on the order of appearance in the dataset.
levels(mtcars$carb) # original levels## [1] "1" "2" "3" "4" "6" "8"fct_inorder(mtcars$carb) # levels based on the order of appearance## [1] 4 4 1 1 2 1 4 2 2 4 4 3 3 3 4 4 4 1 2 1 1 2 2 4 2 1 2 2 4 6 8 2
## Levels: 4 1 2 3 6 8fct_infreq()
This function sorts factor levels based on the frequency of values.
fct_count(mtcars$carb) # this is forcats function as well, count factor level## # A tibble: 6 x 2
## f n
## <fct> <int>
## 1 1 7
## 2 2 10
## 3 3 3
## 4 4 10
## 5 6 1
## 6 8 1levels(mtcars$carb) # original levels## [1] "1" "2" "3" "4" "6" "8"fct_infreq(mtcars$carb) # levels based on the frequency values## [1] 4 4 1 1 2 1 4 2 2 4 4 3 3 3 4 4 4 1 2 1 1 2 2 4 2 1 2 2 4 6 8 2
## Levels: 2 4 1 3 6 8fct_relevel()
This function can be used to change the order manually.
levels(mtcars$carb) # original levels## [1] "1" "2" "3" "4" "6" "8"fct_relevel(mtcars$carb, c("8", "6", "4", "3", "2", "1")) # manually changed new levels## [1] 4 4 1 1 2 1 4 2 2 4 4 3 3 3 4 4 4 1 2 1 1 2 2 4 2 1 2 2 4 6 8 2
## Levels: 8 6 4 3 2 1fct_relevel() can also be used to change one factor level only.
levels(mtcars$carb) # original levels## [1] "1" "2" "3" "4" "6" "8"fct_relevel(mtcars$carb, "8", after = 2) # change level 8 to the third place## [1] 4 4 1 1 2 1 4 2 2 4 4 3 3 3 4 4 4 1 2 1 1 2 2 4 2 1 2 2 4 6 8 2
## Levels: 1 2 8 3 4 6fct_reorder()
This function changes the order based on another variable. Let’s change variable carb’s levels based on value of variable disp.
levels(mtcars$carb) # original levels## [1] "1" "2" "3" "4" "6" "8"fct_reorder(mtcars$carb, mtcars$disp, .fun = sum, .desc = TRUE) # new level based on disp value## [1] 4 4 1 1 2 1 4 2 2 4 4 3 3 3 4 4 4 1 2 1 1 2 2 4 2 1 2 2 4 6 8 2
## Levels: 4 2 1 3 8 6mtcars %>%
group_by(carb) %>%
summarise(sum_disp = sum(disp)) %>%
arrange(desc(sum_disp)) # this is basically what we do with fct_reorder() above## # A tibble: 6 x 2
## carb sum_disp
## <fct> <dbl>
## 1 4 3088.
## 2 2 2082.
## 3 1 940.
## 4 3 827.
## 5 8 301
## 6 6 145Additionally, fct_reorder() can be used with plotting as well.
# Original plot
ggplot(mtcars, aes(x = carb, y = disp)) +
geom_col()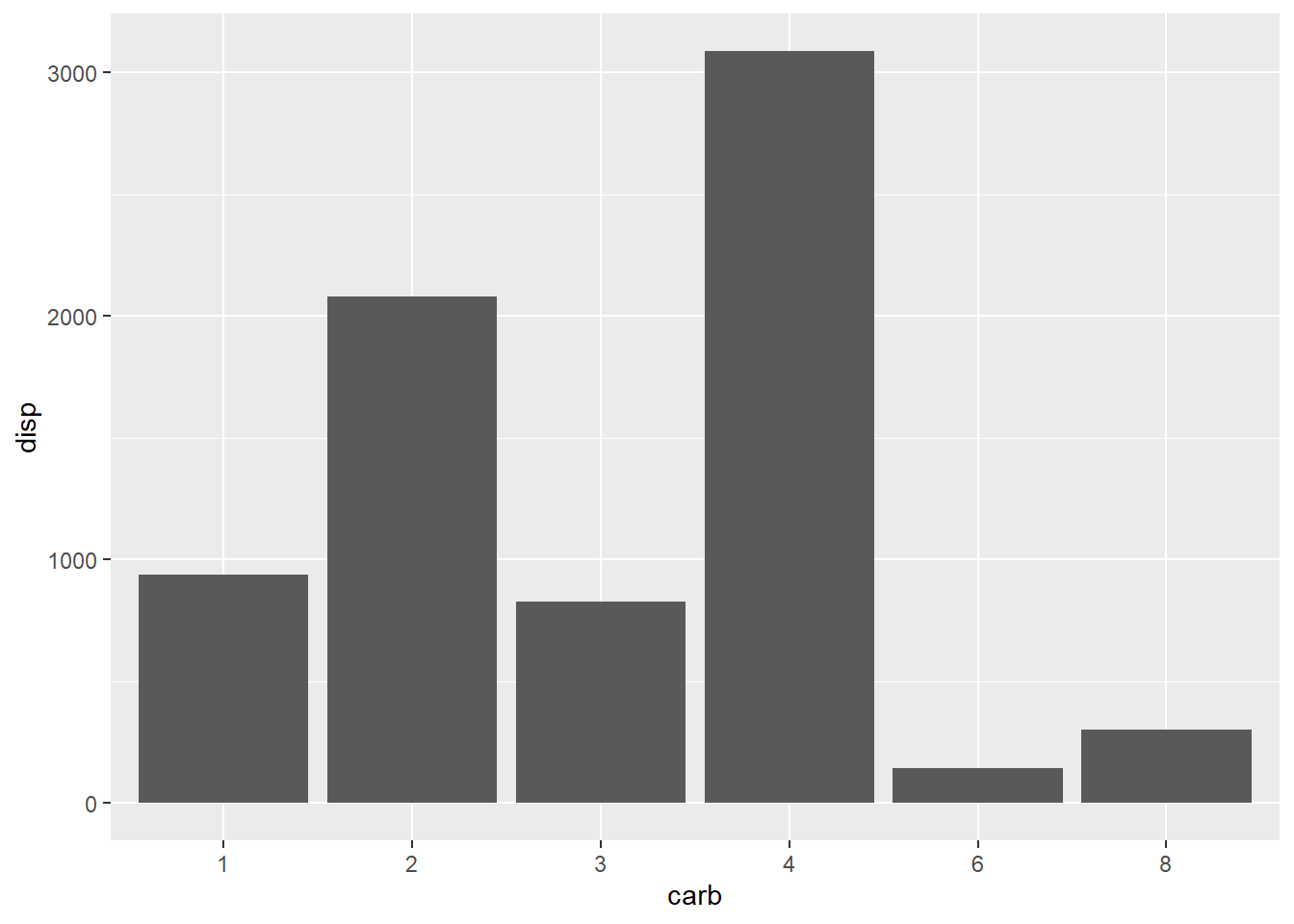
# Plot with changed levels
mtcars %>%
mutate(carb = fct_reorder(carb, disp, .fun = sum, .desc = TRUE)) %>%
ggplot(aes(x = carb, y = disp)) +
geom_col()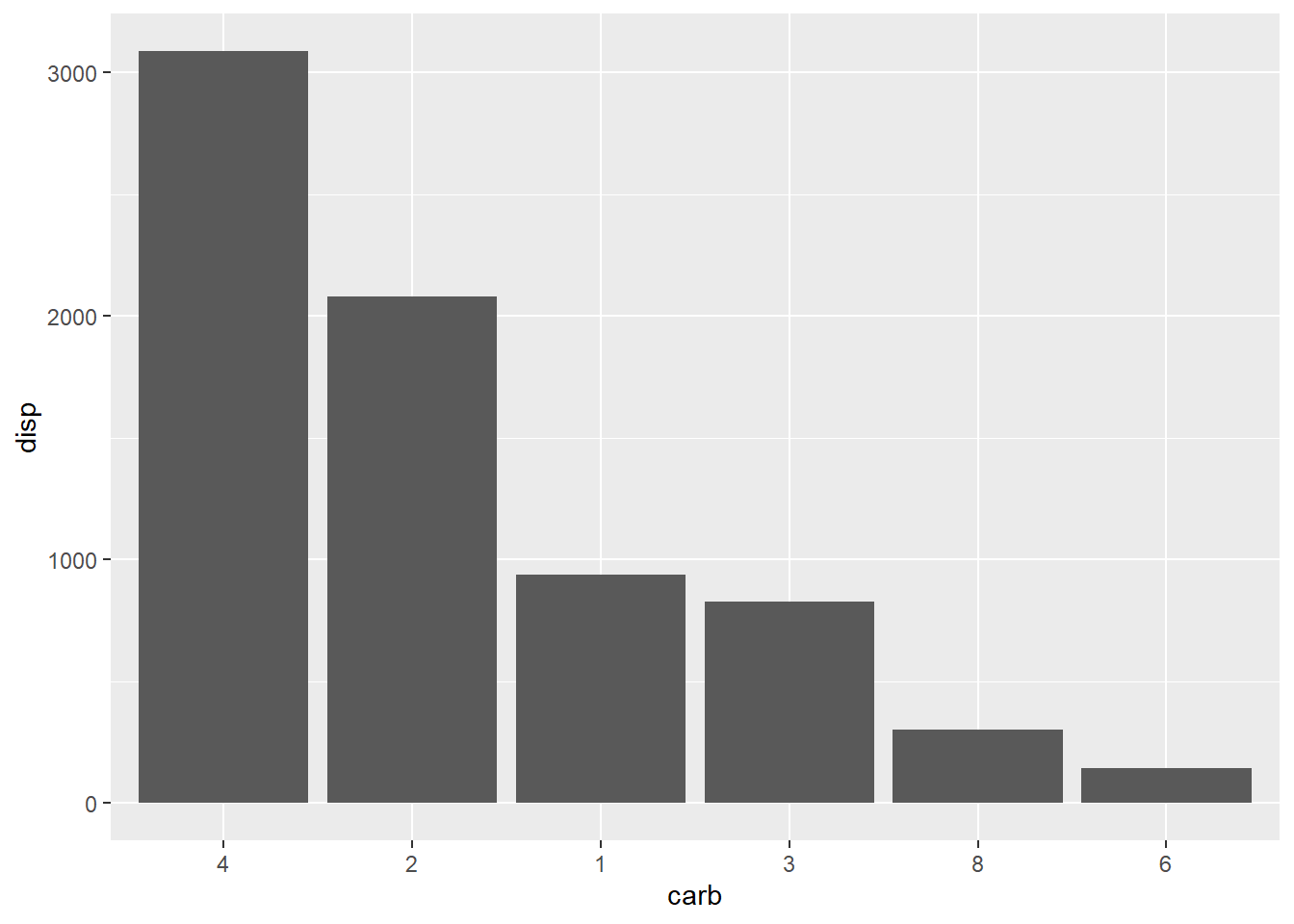
fct_lump()
This function lumps factor levels into other factors. There are 5 variants of this function:
fct_lump()fct_lump_min()fct_lump_n()fct_lump_lowfreq()
The remaining one variant is fct_lump_prop(). It is not in the example below as I do not find it useful at least for my current R coding routine.
fct_lump() automatically lump small frequency factor group into one group.
fct_count(mtcars$carb) # this is forcats function as well, count factor level## # A tibble: 6 x 2
## f n
## <fct> <int>
## 1 1 7
## 2 2 10
## 3 3 3
## 4 4 10
## 5 6 1
## 6 8 1fct_lump(mtcars$carb) %>% fct_count() ## # A tibble: 4 x 2
## f n
## <fct> <int>
## 1 1 7
## 2 2 10
## 3 4 10
## 4 Other 5fct_lump_min() lump factor group into one group based on the given value.
table(fct_lump_min(mtcars$carb, min = 2)) # group 6 and 8 lump into one group##
## 1 2 3 4 Other
## 7 10 3 10 2fct_lump_n() lump all level except for the n most frequent factor groups.
table(fct_lump_n(mtcars$carb, n = 2)) # 2 frequent group only, others in one group##
## 2 4 Other
## 10 10 12fct_lump_lowfreq() lump small frequent groups into one group, while making sure that particular one group is still the smallest.
table(fct_lump_lowfreq(mtcars$carb, other_level = "low")) # group low is still the smallest##
## 1 2 4 low
## 7 10 10 5fct_other()
fct_other() is much like fct_lump(), except we manually choose which factor groups to be combined.
table(fct_other(mtcars$carb, keep = c("8", "6"))) ##
## 6 8 Other
## 1 1 30fct_recode()
This function is used to rename or relabel the factor group.
table(fct_recode(mtcars$carb, hanis = "8")) ##
## 1 2 3 4 6 hanis
## 7 10 3 10 1 1fct_relabel()
fct_relabel() is extremely useful if we want to rename quite a number of factor groups.
table(mtcars$carb) # original groups##
## 1 2 3 4 6 8
## 7 10 3 10 1 1table(fct_relabel(mtcars$carb, ~ c("abu", "ali", "chong", "siti", "krish", "lee"))) # new named groups##
## abu ali chong siti krish lee
## 7 10 3 10 1 1Reference:
https://forcats.tidyverse.org/index.html